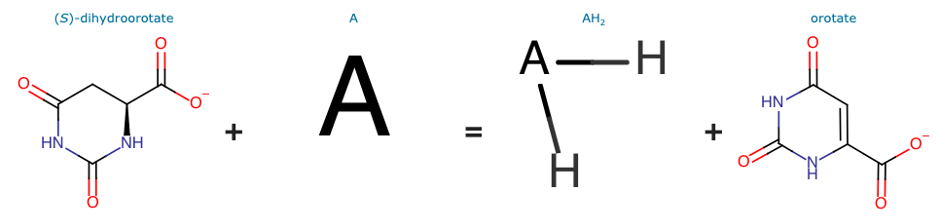Click to Go Back
Examples
Click images to enlarge
Click images to enlarge
Case 1 : A0A1T4KHZ7
Protein: Dihydroorotate dehydrogenase
Gene: pyrD
Organism: Cetobacterium ceti
Motivation: BENZ completes incomplete functional annotation of partially characterized proteins
As of UniProt release 2021_01, TrEMBL A0A1T4KHZ7 contains the protein sequence of a bacterial enzyme, inferred from homology, with an annotation score of 3 out of 5. This enzyme lacks a proper functional characterization but it inherits a partial EC number (1.3.-.-) and a Rhea reaction (RHEA:18073) by the rule UR000081105 derived from the HAMAP rule MF_00224.
Both the partial EC number and the Rhea Reaction aren’t specific enough to clearly explain the catalytic activity performed by the protein. The partial EC number 1.3.-.- indicates a general oxidoreductase that acts on the CH-CH group of the donor. The Rhea reaction represents the conversion of (S)-dihydroorotate in orotate without indicating a specific proton donor and proton acceptor (A and AH2 in the following image).
BENZ in this case can be adopted to refine the functional characterization of the partially annotated protein.
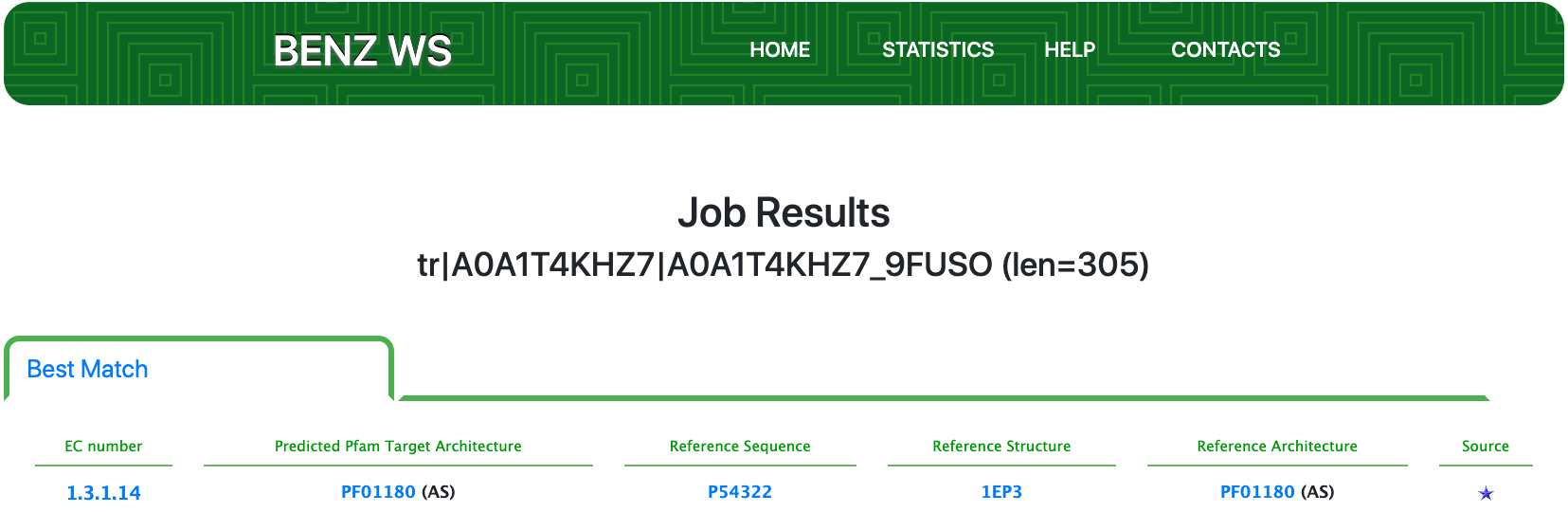
Submitting the sequence in BENZ, it inherits the EC number 1.3.1.14 (Dihydroorotate dehydrogenase (NAD+)) from the matched reference sequence P54322.
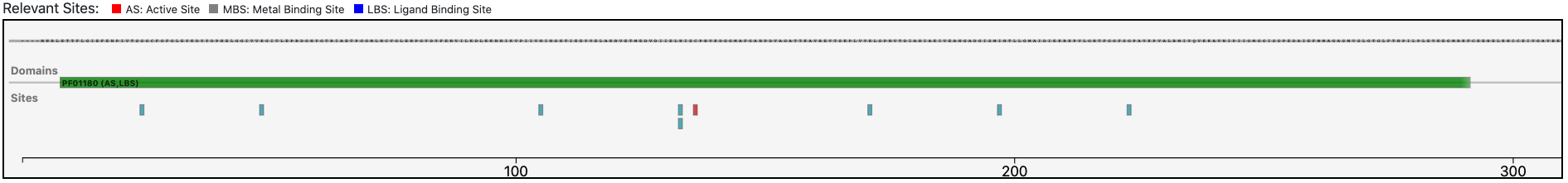
As clearly depicted in the graphical visualization panel, the query protein is predicted with the same Pfam domain architecture as the matched reference.
Furthermore, by inspecting the relevant sites annotated to the reference sequence (P54322) the system allows to verify that relevant sites of the reference are conserved in the query protein. This observation strengthens the functional prediction performed by BENZ.
The functional annotation performed by BENZ is not only compatible with the partial EC number and the Rhea reaction annotated to the query but it allows a four-level EC number prediction.
Case 2 : P07934
Protein: Phosphorylase b kinase gamma catalytic chain, skeletal muscle/heart isoform
Gene: Phkg1
Organism: Mus musculus (Mouse)
Motivation: BENZ can correctly predict polyfunctional proteins matching a specific reference sequence whose relevant sites are conserved in the query protein.
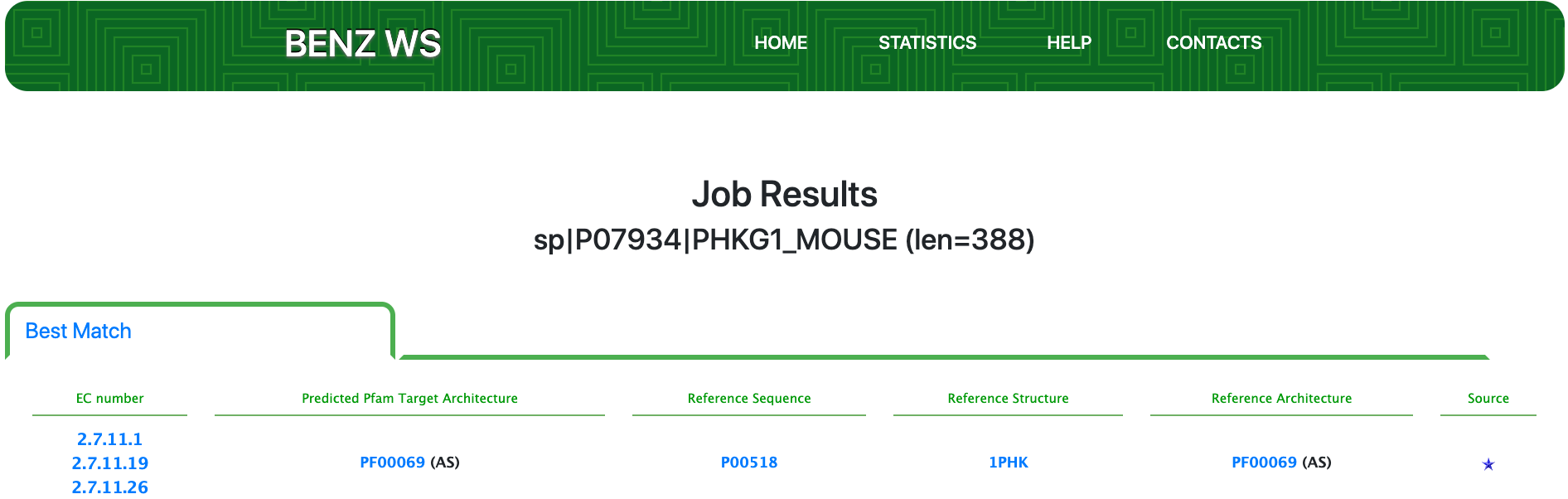
As of UniProt release 2021_01, Swiss-Prot P07934 is a well annotated protein with evidence at the protein level and an annotation score of 5 out of 5. P07934 is the catalytic subunit of the phosphorylase b kinase (PHK) and it’s a polyfunctional enzyme annotated with the EC numbers: 2.7.11.1 (non-specific serine/threonine protein kinase); 2.7.11.19 (phosphorylase kinase) and 2.7.11.26 (tau-protein kinase).
This example shows how BENZ is capable of correctly predicting functions of polyfunctional proteins.
Submitting the sequence in BENZ, it inherits the expected EC numbers 2.7.11.1, 2.7.11.19 and 2.7.11.26 from the matched reference sequence P00518.
The query protein has a single Pfam domain architecture that spans the sequence almost entirely as shown by the graphical visualization panel.
At the same time, the functional prediction of BENZ is corroborated by the conservation in the query protein of the relevant sites annotated in the reference sequence.
Case 3 : Q9P0U3
Protein: Sentrin-specific protease 1
Gene: SENP1
Organism: Homo sapiens (Human)
Motivation: BENZ completes incomplete functional annotation of partially characterized proteins
As of UniProt release 2021_01, Q9P0U3 is a well annotated protein in Swiss-Prot with evidence at the protein level and an annotation score of 5 out of 5. The human enzyme Q9P0U3 catalyzes two essential functions in the SUMO pathway: the hydrolysis of the alpha-linked peptide bond at the C-terminal end of the small ubiquitin-like modifier propeptide (Smt3), leading to the mature form of the protein and the cleavage of an epsilon-linked peptide bond between the C-terminal glycine of the mature SUMO and the lysine epsilon-amino group of the target protein.
Despite the high number of publications linked to the enzyme, Q9P0U3 is annotated in UniProt with a partial EC number: 3.4.22.- (cysteine endopeptidase).
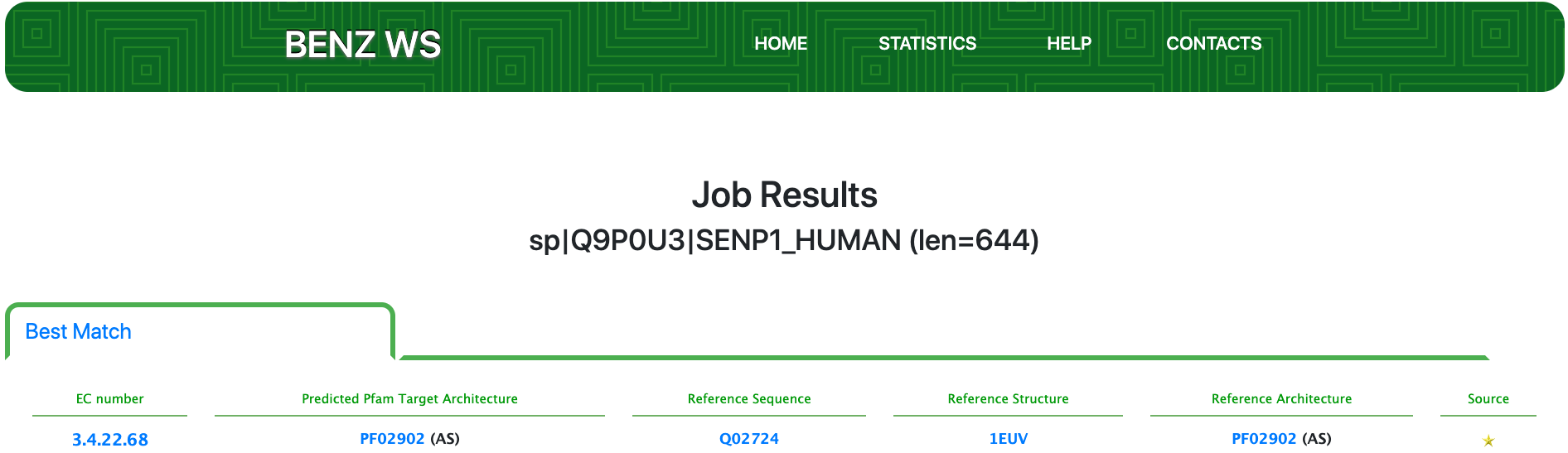
By submitting the sequence to BENZ, it inherits the EC numbers 3.4.22.68 (Ulp1 peptidase) from the matched reference sequence Q02724.
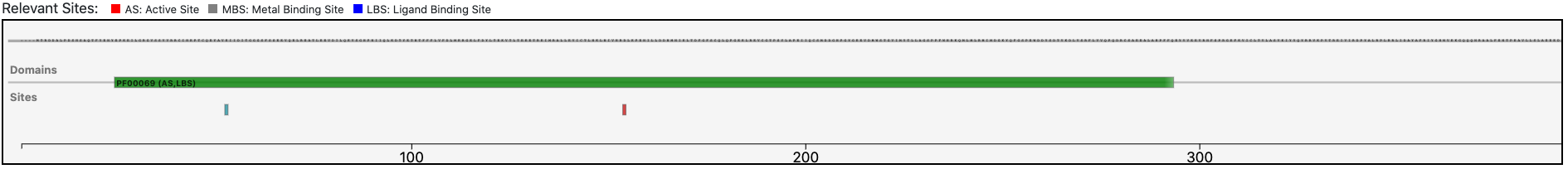
The query protein is predicted having a single Pfam domain architecture located in the C-terminus of the sequence.
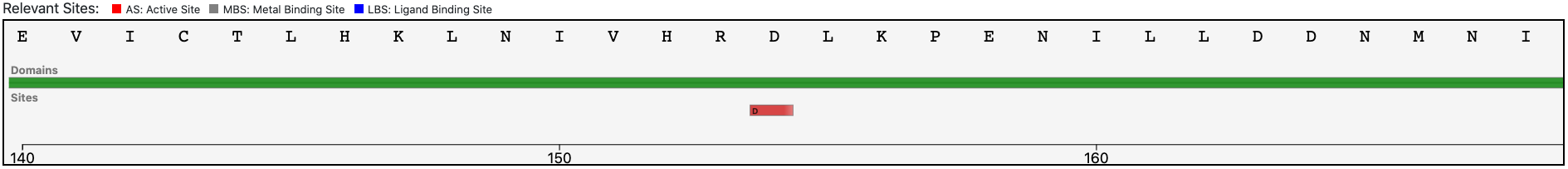
Magnifying the Pfam domain region, we can observe that the active sites annotated in the reference sequence are conserved in the query enzyme, confirming the functional prediction performed by BENZ.
Case 4 : Q76I76
Protein: Protein phosphatase Slingshot homolog 2
Gene: SSH2
Organism: Homo sapiens (Human)
Motivation: BENZ can correctly predict polyfunctional proteins matching a specific reference sequence whose relevant sites are conserved in the query protein.
As of UniProt release 2021_01, Swiss-Prot Q76I76 is a well annotated protein, with evidence at the protein level and an annotation score of 5 out of 5. Q76I76 is a multifunctional phosphatase, which regulates actin filament dynamics dephosphorylating and activating the actin binding/depolymerizing factor cofilin. Q76I76 is annotated with the EC numbers: 3.1.3.16 (protein-serine/threonine phosphatase) and 3.1.3.48 (Protein-tyrosine-phosphatase).
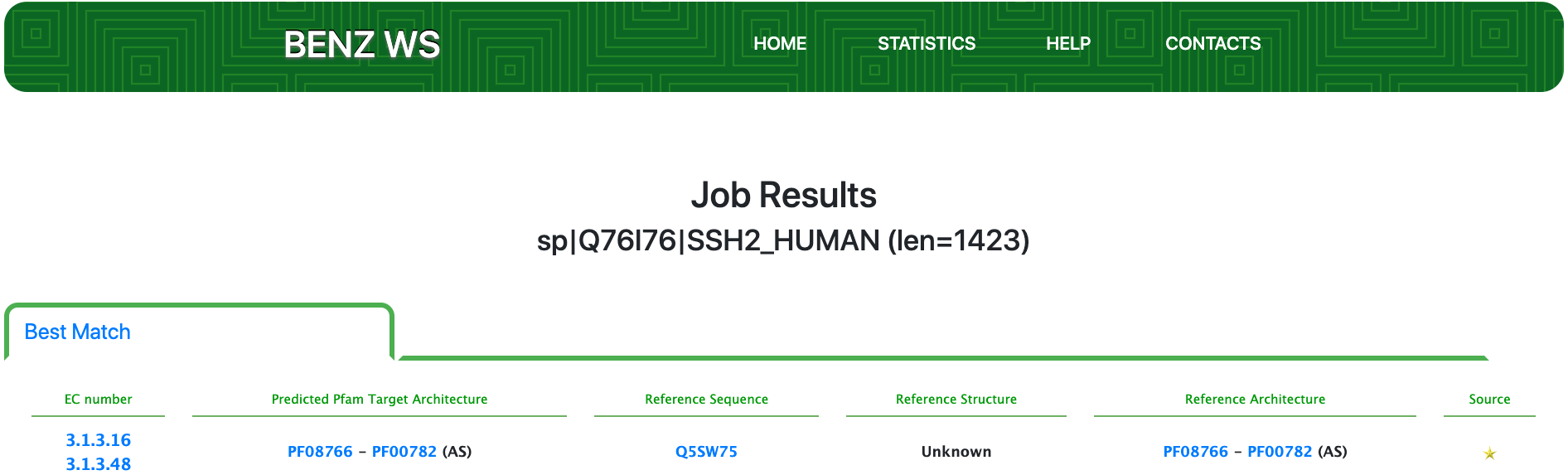
Submitting the sequence to BENZ, it inherits the correct EC numbers 3.1.3.16 and 3.1.3.48 from the matched reference sequence Q5SW75.
The query protein is predicted with a two Pfam domains architecture that covers only a small portion of the entire sequence.
The active site annotated to the reference sequence is conserved in the query enzyme (as depicted below).
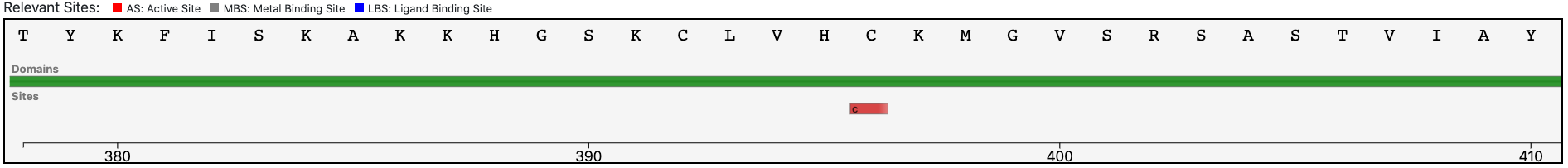
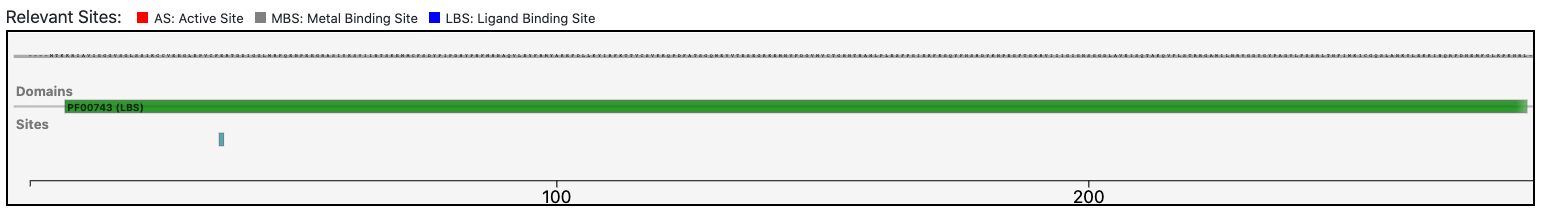
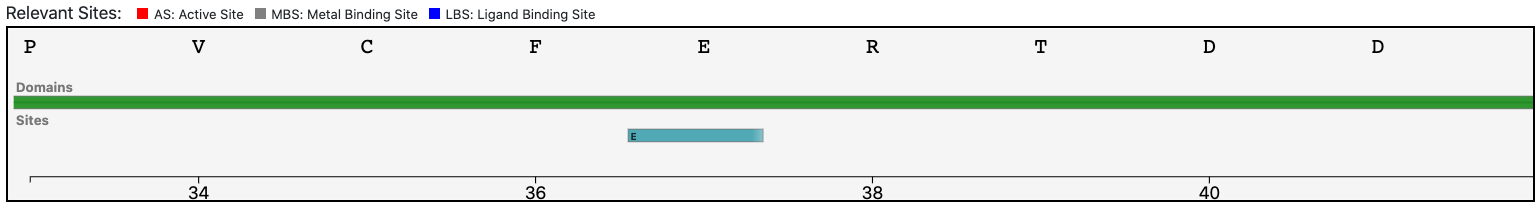
Case 5 : E9PJF3
Protein: Flavin-containing monooxygenase
Gene: FMO5
Organism: Homo sapiens (Human)
Motivation: BENZ completes incomplete functional annotation of partially characterized proteins.
As of UniProt release 2021_01, TrEMBL E9PJF3 contains the protein sequence of a human enzyme with an annotation score of 4 out of 5. This enzyme has a partial EC number 1.-.-.- (Oxidoreductases) from UniRule and two EC numbers, 1.14.13.8 and 1.6.3.1 (flavin-containing monooxygenase) from the ARBA system.
Submitting the sequence in BENZ, it inherits the EC numbers 1.14.13.8 (flavin-containing monooxygenase) and 1.6.3.1 (NAD(P)H oxidase (H2O2-forming)) from the matched reference sequence P49109.
The query protein has a single Pfam domain architecture that covers a large portion of the entire sequence as shown by the graphical visualization panel.
At the same time, the functional prediction of BENZ is corroborated by the conservation in the query protein of the relevant sites annotated in the reference sequence.
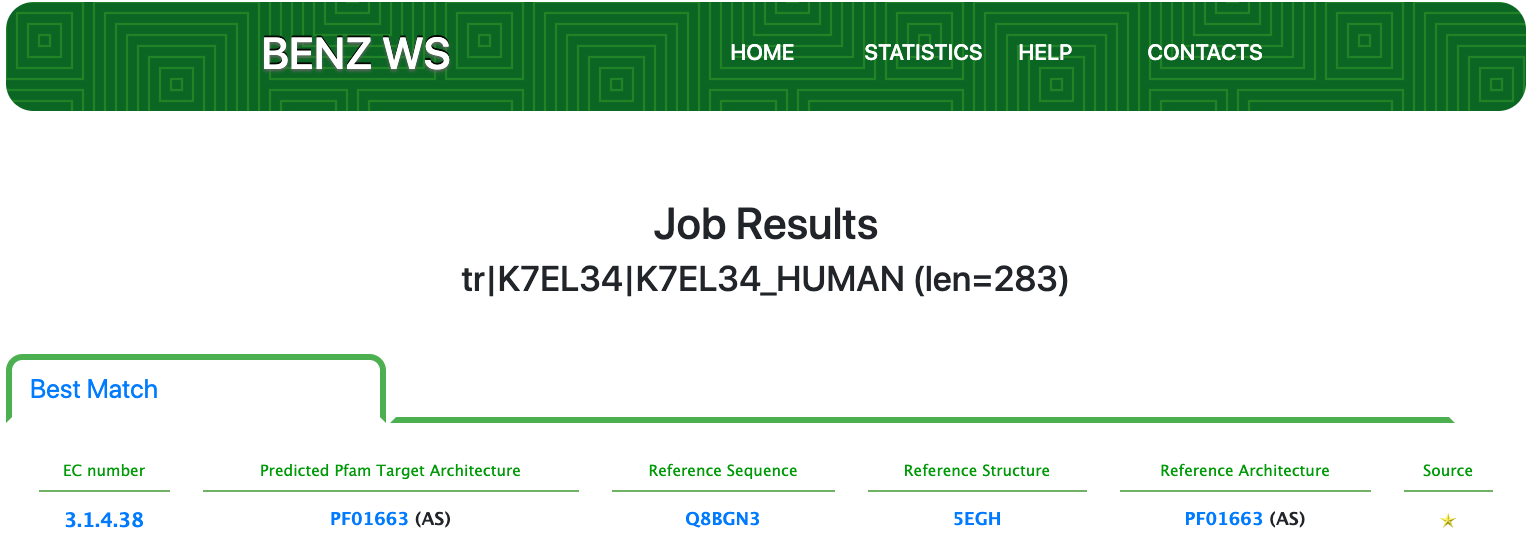
Case 6 : K7EL34
Protein: GPI ethanolamine phosphate transferase 1
Gene: PIGN
Organism: Homo sapiens (Human)
Motivation: BENZ completes incomplete functional annotation of partially characterized proteins.
As of UniProt release 2021_01, TrEMBL K7EL34 is a human enzyme with evidence at the protein level and an annotation score of 3 out of 5. This enzyme is functionally characterized by the UniRule system matching the rule UR001454753. This rule is based on few conditions: matching the PANTHER signature PTHR12250, having a sequence length between 1 and 1200 residues and being expressed in Eukaryota. By means of this rule, K7EL34 is annotated with the partial EC number 2.-.-.- (Transferases).
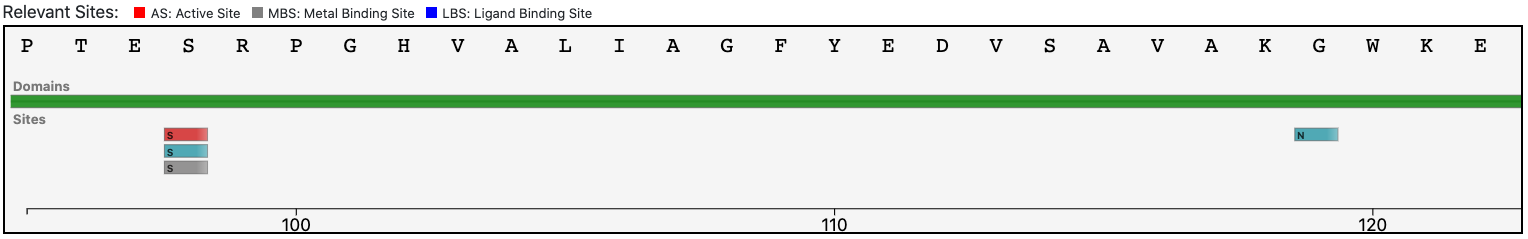
Submitting the sequence in BENZ, it inherits the EC numbers 3.1.4.38 (glycerophosphocholine cholinephosphodiesterase) from the matched reference sequence Q8BGN3.
As clearly depicted in the graphical visualization panel, the query protein is predicted with the same Pfam domain architecture as the matched reference. The same Pfam domain is annotated to the entry in UniProt.
Moreover, by inspecting the relevant sites annotated to the reference sequence (Q8BGN3) BENZ allows to confirm that relevant sites of the reference are conserved in the query protein. This observation suggests that the function predicted by BENZ may be more accurate than the one in UniProtKB.
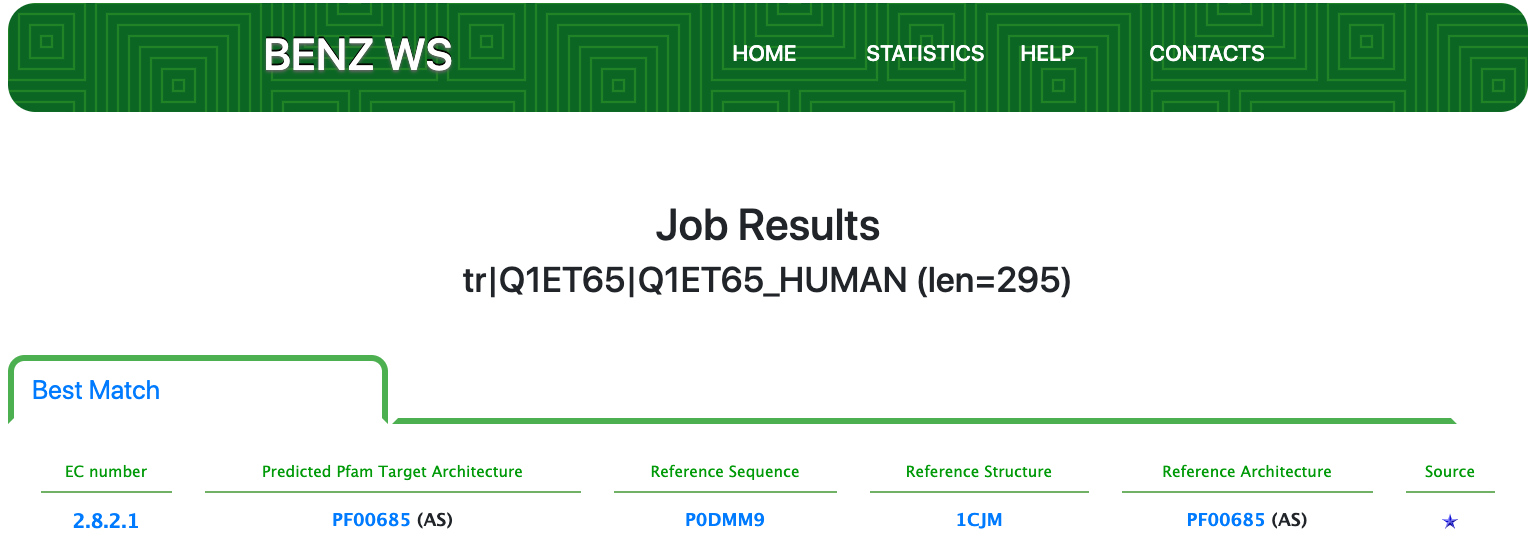
Case 7 : Q1ET65
Protein: Sulfotransferase
Gene: ST1A5
Organism: Homo sapiens (Human)
Motivation: BENZ completes incomplete functional annotation of partially characterized proteins.
As of UniProt release 2021_01, TrEMBL Q1ET65 is a poorly annotated protein with evidence at the transcript level and an annotation score of 1 out of 5. Q1ET65 is annotated with the partial EC number 2.8.2.- (Sulfotransferases) by the UniRule system of annotation.
Submitting the sequence in BENZ, it inherits the EC number 2.8.2.1(aryl sulfotransferase), completing its three-level EC, from the matched reference sequence P0DMM9.
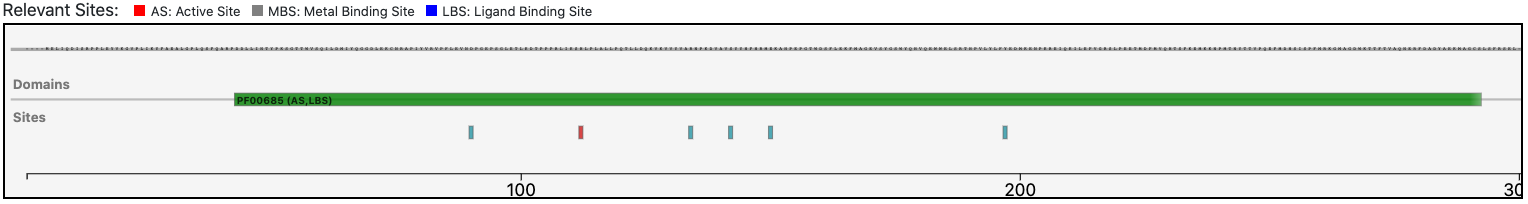
The query protein has a single Pfam domain architecture that spans the sequence almost entirely as shown by the graphical visualization panel.
Magnifying the Pfam domain region, we can observe that the active sites annotated in the reference sequence are conserved in the query enzyme. This observation (as depicted below) confirms the functional prediction performed by BENZ.
Case 8 : Q7Z5G3
Protein: Acetyl-coenzyme A synthetase
Gene: ACSS1
Organism: Homo sapiens (Human)
Motivation: BENZ can confirm functional annotation of partially characterized proteins.
As of UniProt release 2021_01, TrEMBL Q7Z5G3 is a poorly annotated protein with evidence at the transcript level and an annotation score of 2 out of 5. Q7Z5G3 is a polyfunctional enzyme annotated with the EC numbers 6.2.1.1 (acetate—CoA ligase) and 6.2.1.17 (propionate—CoA ligase) by UniRule and ARBA annotation systems.
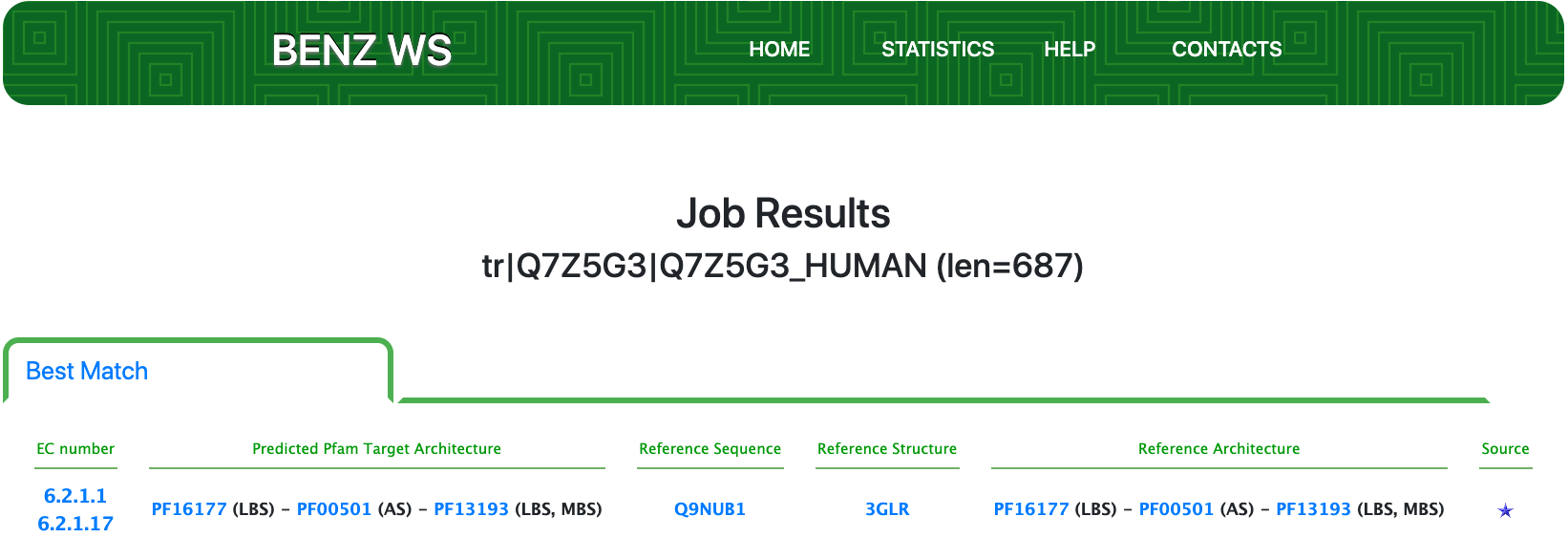
Submitting the sequence to BENZ, it inherits the correct EC numbers 6.2.1.1 and 6.2.1.17 from the matched reference sequence Q9NUB1.
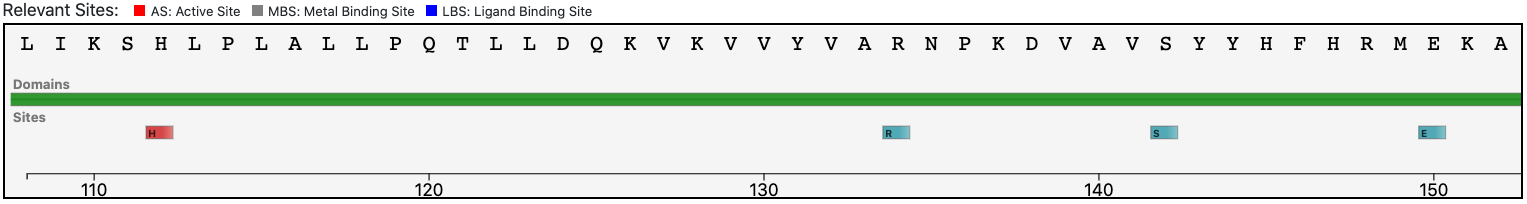
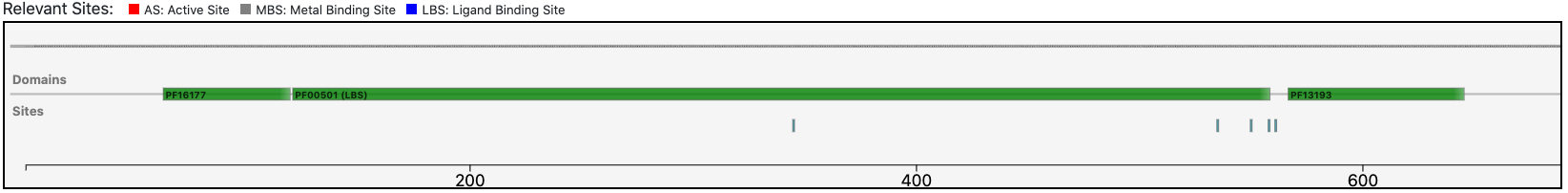
The query protein is predicted with a two Pfam domains architecture that covers almost entirely the sequence.
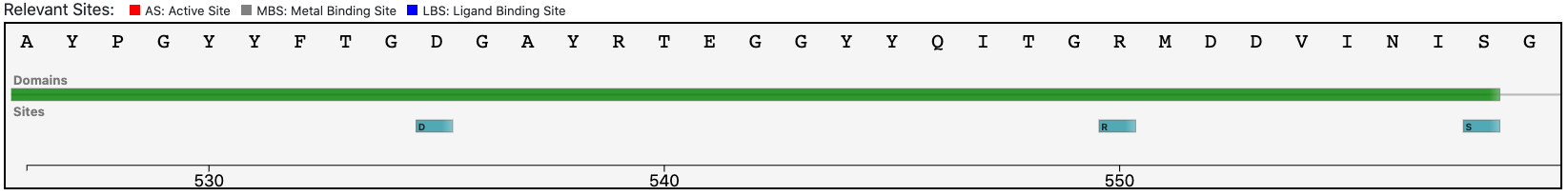
At the same time, the functional prediction of BENZ is supported by the conservation in the query protein of the relevant sites annotated in the reference sequence.
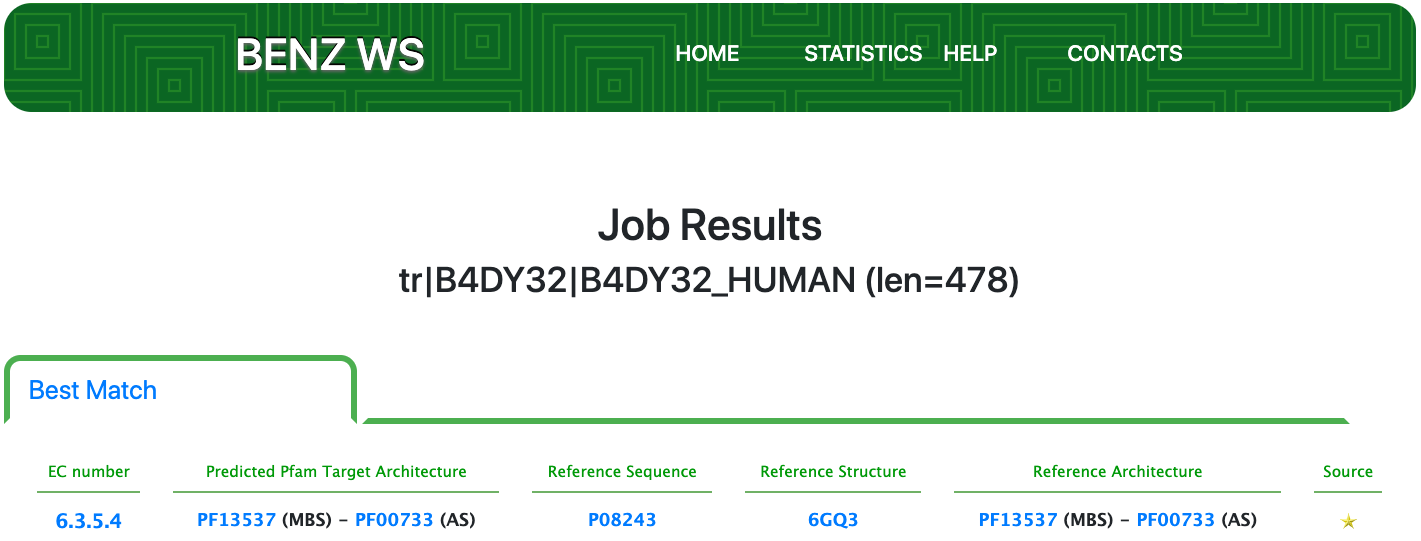
Case 9 : B4DY32
Protein: Asparagine synthetase [glutamine-hydrolyzing]
Gene: N/A
Organism: Homo sapiens (Human)
Motivation: BENZ can confirm functional annotation of poorly characterized proteins.
As of UniProt release 2021_01, TrEMBL B4DY32 is a poorly annotated protein with evidence at the transcript level and an annotation score of 2 out of 5. B4DY32 is annotated with the EC number 6.3.5.4 (asparagine synthase (glutamine-hydrolysing)) by the ARBA rule ARBA00001778.
Submitting the sequence in BENZ, it inherits the expected EC number 6.3.5.4 from the matched reference sequence P08243.
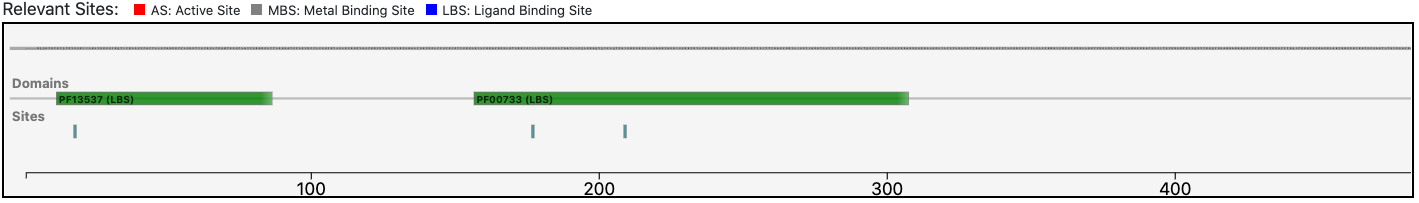
The query protein is predicted having an architecture of two Pfam domains that covers only a portion of the entire sequence.
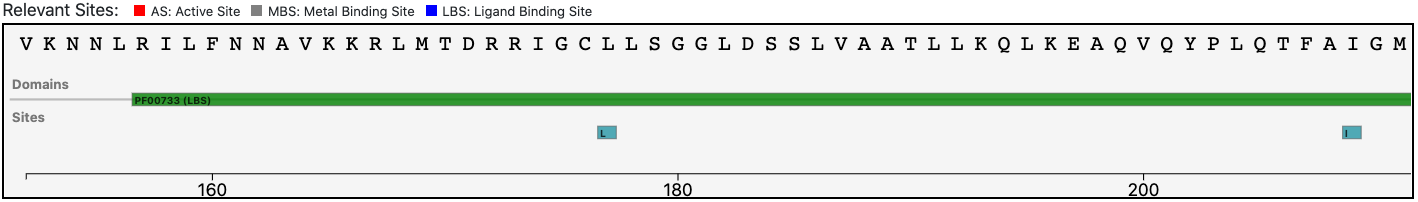
The relevant sites annotated to the reference sequence are conserved in the query enzyme (as depicted below).
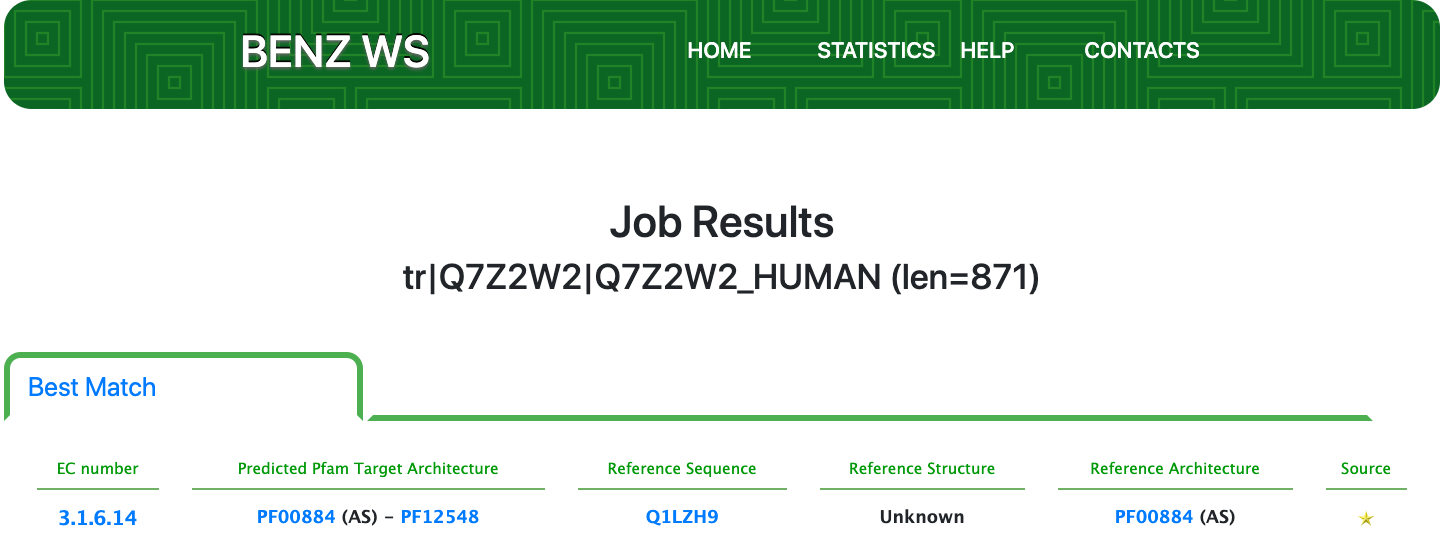
Case 10 : Q7Z2W2
Protein: Extracellular sulfatase
Gene: DKFZp686F13142
Organism: Homo sapiens (Human)
Motivation: BENZ completes incomplete functional annotation of partially characterized proteins
As of UniProt release 2021_01, Q7Z2W2 is a protein in TrEMBL with evidence at the transcript level and an annotation score of 4 out of 5. Q7Z2W2 is a monofunctional enzyme annotated with the partial EC number 3.1.6.- (Sulfuric-ester hydrolases) by UniRule annotation systems.
Submitting the sequence to BENZ, it inherits the EC number 3.1.6.14 (N-acetylglucosamine-6-sulfatase) from the matched reference sequence Q1LZH9.
The query protein is predicted with a two Pfam domains architecture that covers a good part of the sequence.
At the same time, the functional prediction of BENZ is supported by the conservation in the query protein of the relevant sites annotated in the reference sequence.
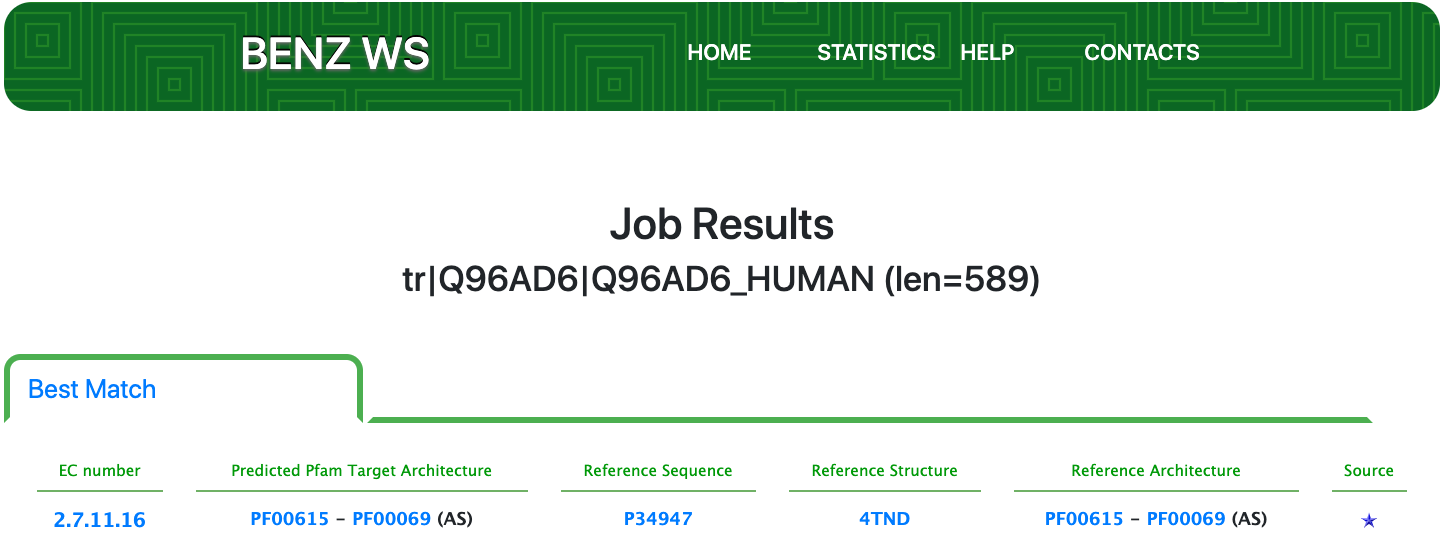
Case 11 : Q96AD6
Protein: G protein-coupled receptor kinase
Gene: GRK6
Organism: Homo sapiens (Human)
Motivation: BENZ completes incomplete functional annotation of partially characterized proteins
As of UniProt release 2021_01, TrEMBL Q96AD6 is a poorly annotated protein with evidence at the transcript level and an annotation score of 2 out of 5. Q96AD6 is annotated with the partial EC number 2.7.11.- (Protein-serine/threonine kinases) by the UniRule UR000000057.
Submitting the sequence in BENZ, it inherits the EC number 2.7.11.16 ([tyrosine 3-monooxygenase] kinase) from the matched reference sequence P34947.
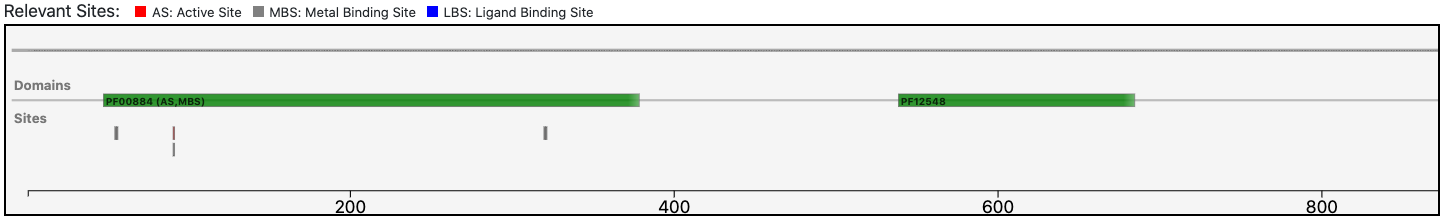
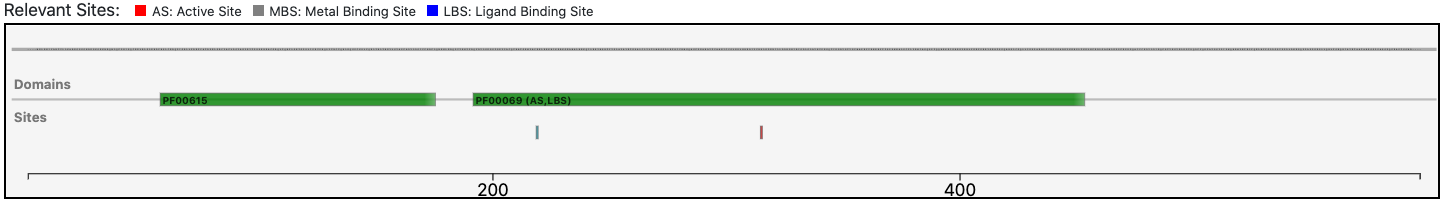
The query protein is predicted having an architecture of two Pfam domains that covers only a portion of the entire sequence.
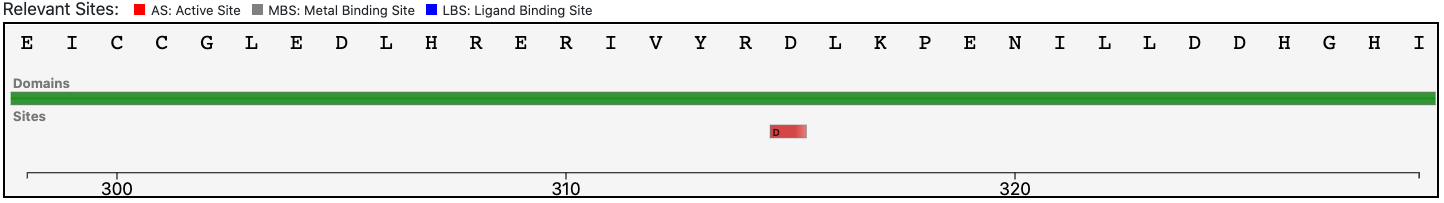
The relevant site annotated to the reference sequence is conserved in the query enzyme (as depicted below).
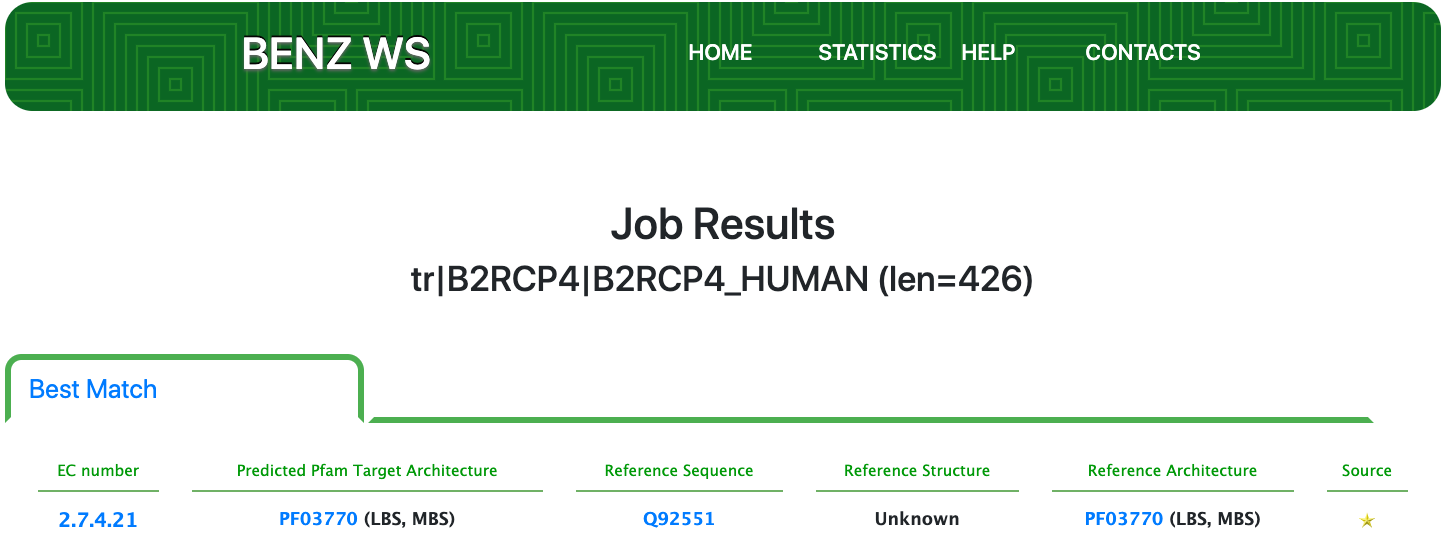
Case 12 : B2RCP4
Protein: Kinase
Gene: IHPK2
Organism: Homo sapiens (Human)
Motivation: BENZ completes incomplete functional annotation of partially characterized proteins
As of UniProt release 2021_01, TrEMBL B2RCP4 is a protein with evidence at the transcript level and an annotation score of 2 out of 5. B2RCP4 is annotated with the partial EC number 2.7.-.- (Transferring phosphorus-containing groups) by the rule UR000342790 mainly based on the match of the PANTHER signature PTHR12400.
Submitting the sequence to BENZ, it inherits the EC number 2.7.4.21 (inositol-hexakisphosphate kinase) from the matched reference sequence Q92551.
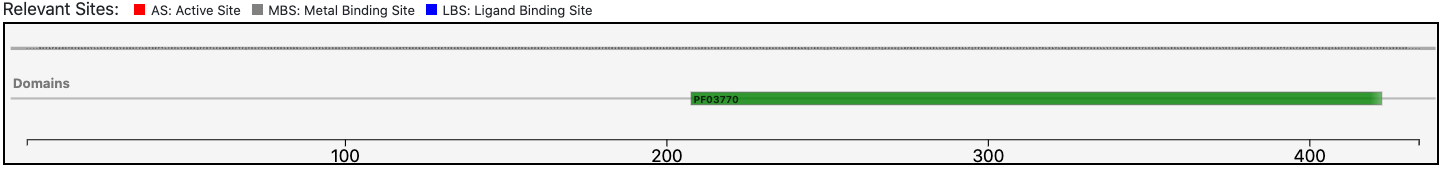
As clearly depicted in the graphical visualization panel, the query protein is predicted with the same Pfam domain architecture as the matched reference.
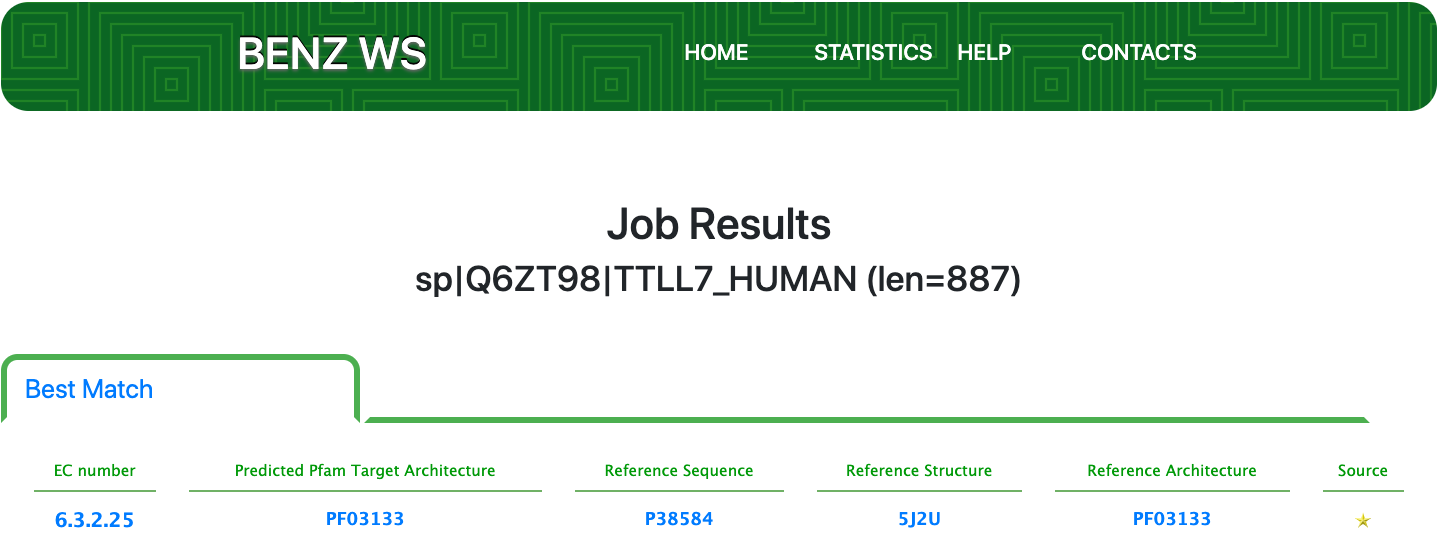
Case 13 : Q6ZT98
Protein: Tubulin polyglutamylase TTLL7
Gene: TTLL7
Organism: Homo sapiens (Human)
Motivation: BENZ completes incomplete functional annotation of partially characterized proteins
As of UniProt release 2021_01, Swiss-Prot Q6ZT98 is a protein with evidence at the protein level and an annotation score of 5 out of 5. Despite the good level of annotation, Q6ZT98 is annotated in UniProt with the partial EC number 6.-.-.- (Ligases).
Submitting the sequence to BENZ, it inherits the EC number 6.3.2.25 (tubulin—tyrosine ligase) from the matched reference sequence P38584.
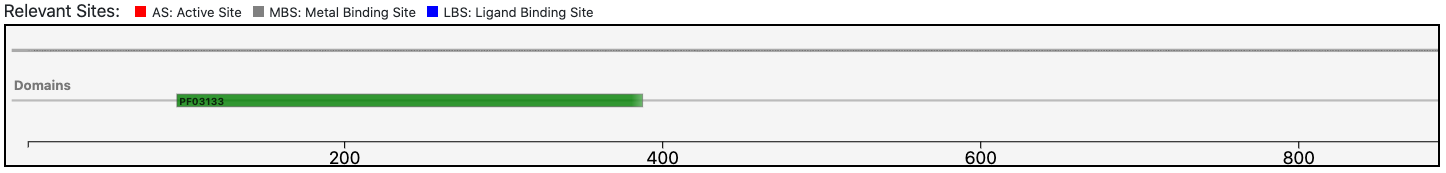
As clearly depicted in the graphical visualization panel, the query protein is predicted with the same Pfam domain architecture as the matched reference.
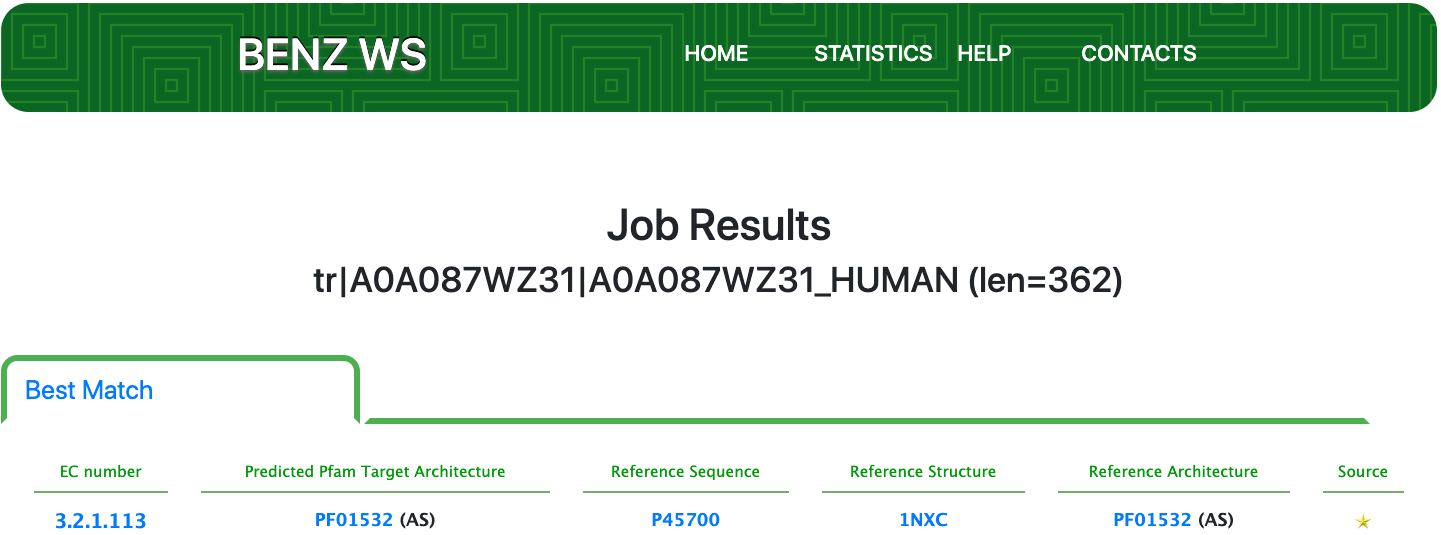
Case 14 : A0A087WZ31
Protein: alpha-1,2-Mannosidase
Gene: MAN1C1
Organism: Homo sapiens (Human)
Motivation: BENZ completes incomplete functional annotation of partially characterized proteins
As of UniProt release 2021_01, TrEMBL A0A087WZ31contains the protein sequence of a human enzyme with an annotation score of 2 out of 5. This enzyme has a partial EC number 3.2.1.- (Glycosidases, i.e. enzymes that hydrolyse O- and S-glycosyl compounds) from UniRule (UR000129775).
Submitting the sequence to BENZ, it inherits the EC number 3.2.1.113 (mannosyl-oligosaccharide 1,2-α-mannosidase) from the matched reference sequence P45700.
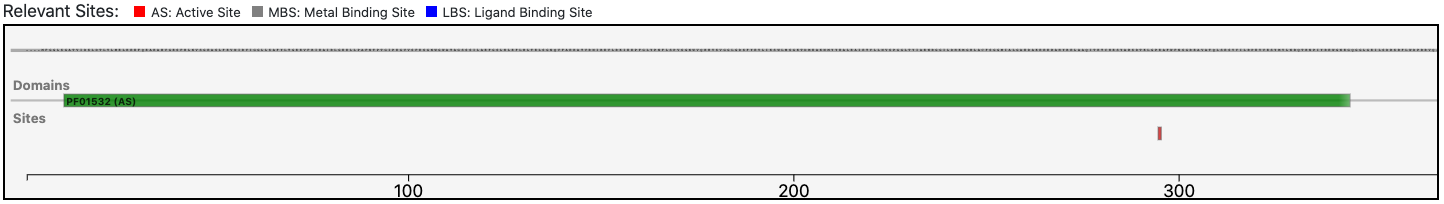
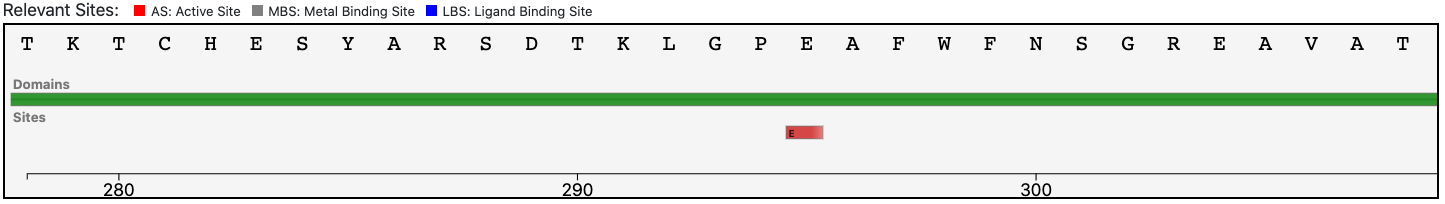
The query protein has a single Pfam domain architecture that covers almost entirely the sequence as shown by the graphical visualization panel.
At the same time, the functional prediction of BENZ is corroborated by the conservation in the query protein of the relevant sites annotated in the reference sequence.