STATISTICS
BENZ WS Content
| EC 1 | EC 2 | EC 3 | EC 4 | EC 5 | EC 6 | EC 7 | Total | |
|---|---|---|---|---|---|---|---|---|
| EC numbers + | 1,437 | 1,550 | 1,034 | 595 | 254 | 189 | 77 | 5,136 |
| Cluster HMM | 1,758 | 5,116 | 3,755 | 1,006 | 637 | 636 | 188 | 12,612 |
| Cluster HMM GOLD | 1,326 | 4,315 | 3,190 | 800 | 497 | 196 | 218 | 10,547 |
| Cluster HMM BLUE | 432 | 801 | 565 | 206 | 140 | 140 | 70 | 2,065 |
| Ref Seq * | 2,752 (390) | 6,455 (990) | 4,582 (729) | 1,348 (324) | 842 (145) | 883 (105) | 405 (14) | 16,593 (2,023) |
| Ref Str * | 1,230 (152) | 2,252 (253) | 2,100 (213) | 625 (110) | 333 (41) | 287 (22) | 149 (5) | 6,798 (618) |
| PFAM ° | 682 (429) | 1,923 (769) | 1,672 (711) | 463 (321) | 294 (190) | 276 (133) | 143 (50) | 4,158 (1,758) |
| KEGG ID ^ | 2,390 | 5,908 | 3,770 | 1,185 | 799 | 894 | 343 | 14,745 |
| KEGG Pathway ^ | 2,758 | 4,812 | 2,628 | 1,972 | 952 | 1,266 | 317 | 9,382 |
| Organisms ** |
|
|
|
|
|
|
|
|
+ Four-level EC numbers distributed according to the 7 EC classes: EC1-Oxidoreductases, EC2-Transferases, EC3-Hydrolases, EC4-Lyases,EC5-Isomerases, EC6-Ligases, EC7-Translocases.
* Ref Seq and Ref Str: number of reference sequences, and reference sequence with structure, respectively; number of polyfunctional enzymes are within brackets.
° Pfam: models from Pfam (https://pfam.xfam.org) ; within brackets Pfams, where relevant sites (active, metal, ligand binding site) are annotated.
^ KEGG ID: from UniProt annotation; KEGG pathway: from https://www.genome.jp/kegg/.
** number of organisms detailed for each kingdom. Arc: Archaea; Bac: Bacteria; Euk: Eukaryota; Oth: Others; Vir: Viruses. Unk: unknown. Annotation source: UniProt. Grand Total: 891.
* Ref Seq and Ref Str: number of reference sequences, and reference sequence with structure, respectively; number of polyfunctional enzymes are within brackets.
° Pfam: models from Pfam (https://pfam.xfam.org) ; within brackets Pfams, where relevant sites (active, metal, ligand binding site) are annotated.
^ KEGG ID: from UniProt annotation; KEGG pathway: from https://www.genome.jp/kegg/.
** number of organisms detailed for each kingdom. Arc: Archaea; Bac: Bacteria; Euk: Eukaryota; Oth: Others; Vir: Viruses. Unk: unknown. Annotation source: UniProt. Grand Total: 891.
Polyfunctional☨︎ enzymes
| EC 1 | EC 2 | EC 3 | EC 4 | EC 5 | EC 6 | EC 7 | Total | |
|---|---|---|---|---|---|---|---|---|
| EC numbers + | 384 | 475 | 311 | 255 | 84 | 70 | 10 | 1,589 |
| Cluster HMM | 261 | 736 | 564 | 227 | 104 | 72 | 11 | 1,485 |
| Cluster HMM GOLD | 135 | 472 | 376 | 127 | 50 | 36 | 6 | 907 |
| Cluster HMM BLUE | 126 | 264 | 188 | 100 | 54 | 36 | 5 | 578 |
| Ref Seq * | 390 | 990 | 729 | 324 | 145 | 105 | 14 | 2,023 |
| Ref Str * | 152 | 253 | 213 | 110 | 41 | 22 | 5 | 618 |
| PFAM ° | 254 (207) | 686 (389) | 532 (302) | 209 (169) | 112 (90) | 97 (61) | 8 (5) | 1,156 (725) |
| KEGG ID ^ | 346 | 841 | 598 | 281 | 133 | 109 | 12 | 1,776 |
| KEGG Pathway ^ | 800 | 1,378 | 797 | 709 | 342 | 292 | 14 | 2,627 |
| Organisms ** |
|
|
|
|
|
|
|
|
☨ For polyfunctional enzymes, four-level EC codes are counted independently for the seven enzymatic classes.
+ Four-level EC numbers distributed according to the 7 EC classes: EC1-Oxidoreductases, EC2-Transferases, EC3-Hydrolases, EC4-Lyases,EC5-Isomerases, EC6-Ligases, EC7-Translocases.
* Ref Seq and Ref Str: number of reference sequences, and reference sequence with structure, respectively; number of polyfunctional enzymes are within brackets.
° Pfam: models from Pfam (https://pfam.xfam.org) ; within brackets Pfams, where relevant sites (active, metal, ligand binding site) are annotated.
^ KEGG ID: from UniProt annotation; KEGG pathway: from https://www.genome.jp/kegg/.
** number of organisms detailed for each kingdom. Arc: Archaea; Bac: Bacteria; Euk: Eukaryota; Oth: Others; Vir: Viruses. Unk: unknown. Annotation source: UniProt. Grand Total: 384.
+ Four-level EC numbers distributed according to the 7 EC classes: EC1-Oxidoreductases, EC2-Transferases, EC3-Hydrolases, EC4-Lyases,EC5-Isomerases, EC6-Ligases, EC7-Translocases.
* Ref Seq and Ref Str: number of reference sequences, and reference sequence with structure, respectively; number of polyfunctional enzymes are within brackets.
° Pfam: models from Pfam (https://pfam.xfam.org) ; within brackets Pfams, where relevant sites (active, metal, ligand binding site) are annotated.
^ KEGG ID: from UniProt annotation; KEGG pathway: from https://www.genome.jp/kegg/.
** number of organisms detailed for each kingdom. Arc: Archaea; Bac: Bacteria; Euk: Eukaryota; Oth: Others; Vir: Viruses. Unk: unknown. Annotation source: UniProt. Grand Total: 384.
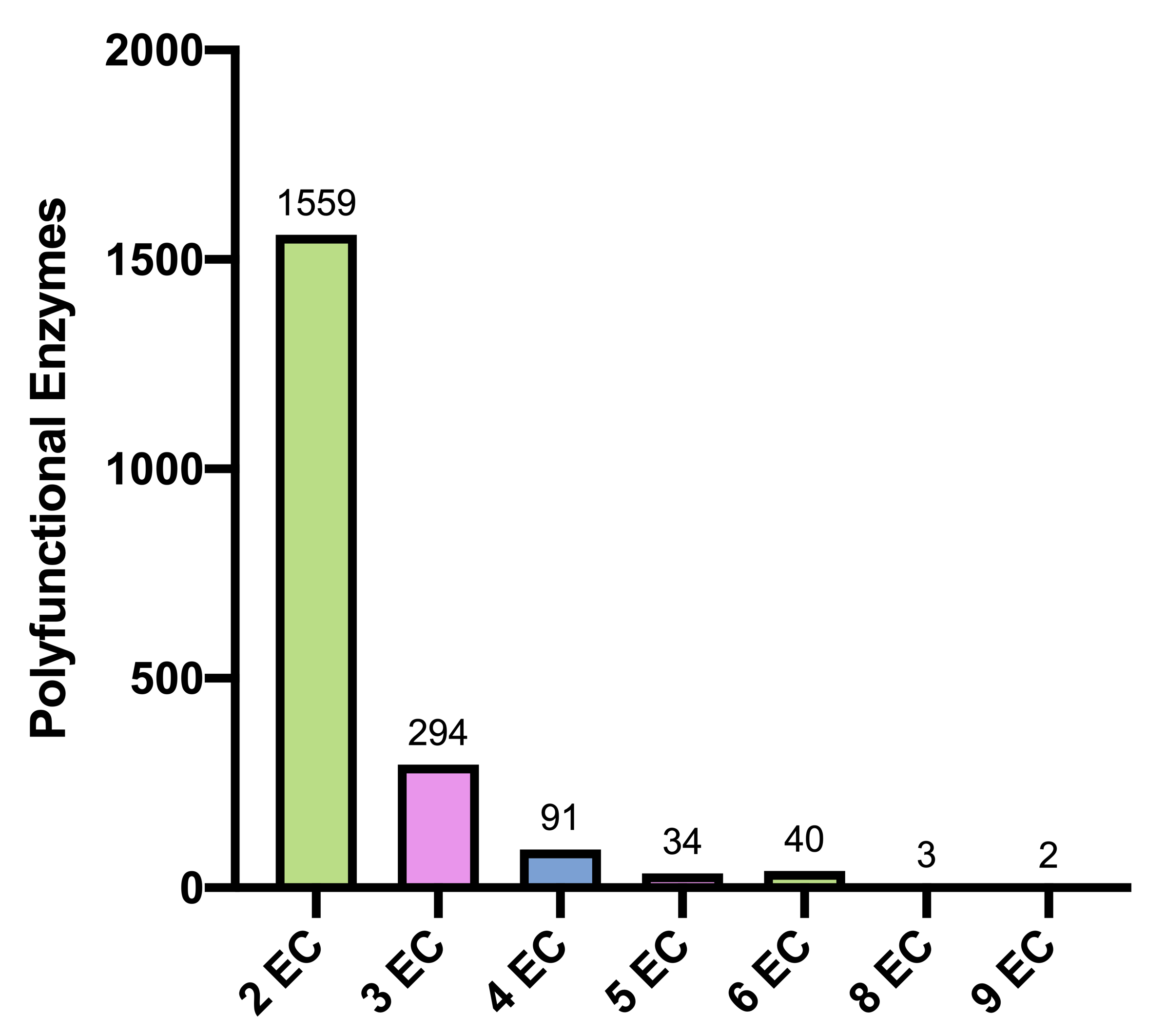
Fig.1S Reference polyfunctional enzymes are grouped according to the associated number of four-level ECs (Grand Total: 2,023).